DEVMAP Project
This is a collaboration project with INRA - Rennes. The main purpose of the project is to study the complex interactions between the evolution of insect populations and environmental factors affecting their functions. The project focus on Carabidae, one of the most common and beneficial insects in France. The first step is the digital imaging of Carabidae specimens. Then, the images will be studied by applying the analysis methods. The methods which have been used to study are based on the morphometry analysis and deep learning.
MAELab Framework
MAELab is a framework in image processing which provides the functions for automatic landmarking on biological images, specific beetle's anatomical species. Besides, it has also implemented the commonly used functions in image processing domain, such as segmentation, binary operations, or filter operations. MAELab is written in C++ and used LibJpeg to read the images. Currently, MAELab is distributed under two modes with and without graphics user interface (GUI). In GUI mode, the interface is implemented by using Qt Framework. The GUI version is free to download from Github.
Videos
GUI and basic functions presentation
Estimate landmarks on right mandible
Publications
Van Linh LE, Marie BEURTON-AIMAR, Adrien KRAHENBUHL, Nicolas PARISEY, MAELab: a framework to automatize landmark estimation , International Conference in Central Europe on Computer Graphics, Visualization and Computer Vision, 2017 (WSCG-17)
Van Linh LE, Marie BEURTON-AIMAR, Adrien KRAHENBUHL, Nicolas PARISEY, SIFT descriptor to set landmarks on biological images , XXVIe Colloque GRETSI, 2017
LE Van Linh, BEURTON-AIMAR Marie, SALMON Jean-Pierre, ALEXIA Marie, PARISEY Nicolas, Estimating landmarks on 2D images of beetle mandibles , International Conference in Central Europe on Computer Graphics, Visualization and Computer Vision, 2015 (WSCG-15)
DEVMAP with Deep Learning
The first episode has used the image processing techniques to estimate the landmarks. These techniques have provided good estimation on left and right mandible which are considered as easy to segment the interested object. But, it seems that these techniques are not suitable for the cases of noise in segmentation step. In another scenario, we have applied another technique to predict the landmarks: deep learning. We have proposed a Convolutional Neural Network which composed from three elementary blocks followed by 3 fully connected layers. Each elementary block consists of a convolutional layer, a maximum pooling layer and a dropout layer. After training, the proposed model has enabled to predict the landmarks on all parts beetles. All the implementation of model have been provided on Github.
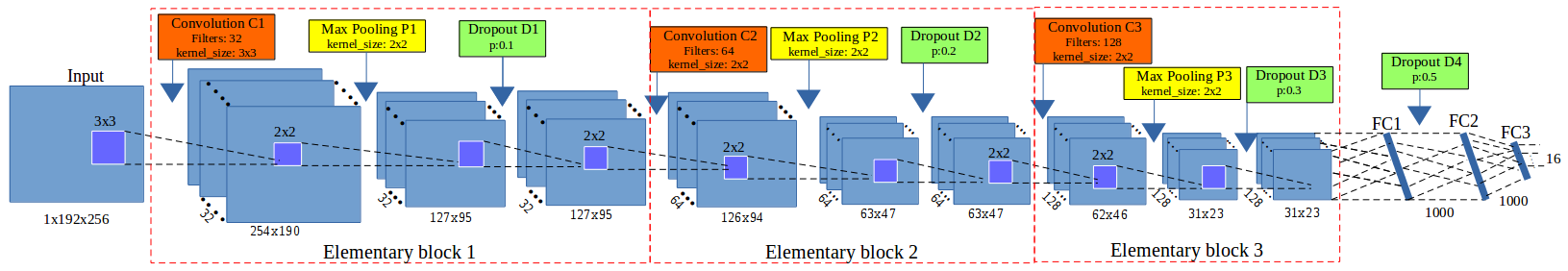
Besides the model, we have implemented a tool to test the trained model on an un-seen image. The program input a new image and trained model, then it will output the predicted landmarks. Notice that we need to choose correctly the model and test image, e.g. we can not use "elytre" model to predict the landmarks on "pronotum" image (see videos below). All the source codes are available on Github.
Videos
Landmarks prediction on pronotum anatomical
Landmarks prediction on elytra anatomical
Landmarks prediction on head anatomical
Publications
Van Linh LE, Marie BEURTON-AIMAR, Akka ZEMMARI, Nicolas PARISEY, Automated morphometrics using deep neural networks: a case study on a beneficial insect species, 10th Symposium National de Morphometrie et Evolution des Formes (SMEF), 2018 (SMEF-18)
Van Linh LE, Marie BEURTON-AIMAR, Akka ZEMMARI, Nicolas PARISEY, Towards landmarks prediction with Deep Network, 9th International Conference on Pattern Recognition Systems, 2018 (ICPRS-18)
Van Linh LE, Marie BEURTON-AIMAR, Akka ZEMMARI, Nicolas PARISEY, Landmarks detection by applying Deep networks, 1st International Conference on Multimedia Analysis and Pattern Recognition, 2018 (MAPR - 2018)


